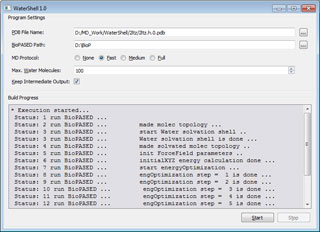
General layout of WaterShell program.
To account for molecular effects, such as van der Waals interactions, hydrogen bonds and water bridges, but keeping computational cost at the minimum, one should use a reliable tool for searching of tightly bound water molecules in an adjacent solvent layer. These molecules can be modeled explicitly, and computational efficiency depends on careful minimization of molecule count. At the first step a molecule of a biololymer is solvated, i.e. surrounded by a layer of arbitrary oriented water molecules within 4.5–5.0 Å. At the second step the derived structure is optimized using molecular dynamics method implemented in BISON package, gradually heating the system to 300 K and modeling in equilibrium state for several nanoseconds. Moreover the structure of the biopolymer is positionally restrained (0.25 kcal/mol•Å2), because the point is to relax and optimize solvent only. Solvent is, on the contrary, not restrained, and water molecules feel free to sample the space for local energy minimums. The resulting trajectories are analyzed at the third step: water molecules forming hydrogen bonds with two, three or more donors and acceptors of the biopolymer, are discriminated. Water list is arranged accoring to number of atoms in a bridge and to occurence along the trajectory, and then at the fourth step, a model of the biopolymer containing these explicitly associated, i.e. "tightly bound", water molecules is built. The derived model can then be implicitly solvated, and contribution of associated water molecules is considered explicitly within the bounds of the selected explicit solvent model (TIP3P, TIP4P and so on).
Program with a graphical user interface WaterShell helps to automate this process. User can select a maximum number of explicit water molecules kept within the model (this amount generally never exceeds 100) and optimization protocol length. Besides there is an option to keep intermediate computation results that are removed by default since they can occupy a huge amount of disk space. The program supports Microsoft Windows operating system family, the language of its interface is English. WaterShell depends on BISON package to run (BioPASED program).
If you use the WaterShell program in any work distributed or published, please include the following reference:
Popov A.V., Vorob'ev Yu.N. 2010. GUI-BioPASED program for molecular dynamics modelling of biopolymers with a graphical user interface. Molecular Biology (Moscow). 44. 735–742.
Links on the topic: