General layout of MDTRA program.
Analysis of molecular dynamics trajectories is a resource-intensive task, and taking into account its present sizes, it is often a rate-limiting step of computer assisted modelling. A lot of programs has been developed to automate and speed up this task, but most of them work in a text mode and require proper scripts to be written. Existing graphical solutions have limited capabilities of result visualization and rely on third-party plotting software, or are not flexible enough in specifying parameters to analyze, or in reading different trajectory formats. The Molecular Dynamics Trajectory Analyzer (MDTRA) program has been developed to analyze trajectories in PDB format, particularly - trajectories output by BioPASED molecular dynamics program. Other trajectory formats, for example, DCD files output by NAMD program, have to be converted into a set of PDB files. It can be performed using VMD program, which creates a single huge PDB file, containing all the snapshots; then that file have to be split into individual files by simple shell scripts distributed with MDTRA.
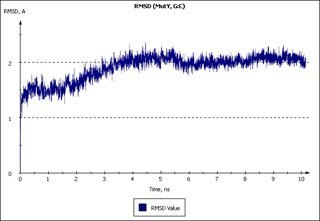
Plot image of RMSD values calculated along a 10 ns trajectory, exported by MDTRA.
MDTRA program analyzes different geometrical parameters of a model (root-mean-square deviation, distances, angles, dihedral angles, angles between planes, etc.) for a given atoms or atomic sets, calculates statistical parameters for the values along the trajectory (mean values, standard deviation, dispersion, median, etc.), linear correlation between trajectories, and builds graphs. Flexible graph options (colors, scalee units, anti-aliasing, legend) along with an automation of some building aspects (for example, reference axes scales) enable fast building of images of high quality and to export them to the one of several common graphics formats to include into a presentation or an article. There are also tools that facilitate a search of significant changes of a structure along the particular trajectory (building a 2D-RMSD plot) or between two trajectories (distance search tool). Some optimizations for multicore processors and SIMD instructions are used to speed up a calculation process. Data calculated is saved to a project file of internal format, and then can be worked with (viewed, exported to text files, shown or hidden on a plot) without the trajectories themselves. In addition to existing tools, MDTRA integrates with PDB file viewers (RasMol, VMD) in order to inspect a structure of particular trajectory elements visually.
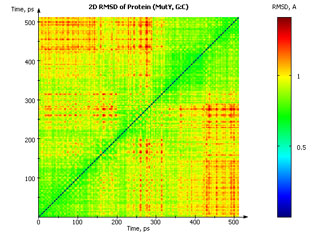
Two-dimensional RMSD plot of values calculated along a 500 ps trajectory fragment, exported by MDTRA (2D-RMSD Tool). Each point at the two-dimensional plot corresponds to the RMSD between two certain trajectory elements, and its value is encoded into a color.
The program supports Microsoft Windows operating system family (starting from Windows XP SP2) and Linux in X-windows mode, the language of its interface is English.
MDTRA can be downloaded from the web site for free. If you use this program in any work distributed or published, please include the following reference:
Popov, A.V., Vorobjev, Y.N., Zharkov, D.O. MDTRA: A molecular dynamics trajectory analyzer with a graphical user interface. J. Comput. Chem. 2013, 34, 319-325. DOI: 10.1002/jcc.23135.
Links on the topic: